Microscopy image segmentation via point and shape regularized data synthesis
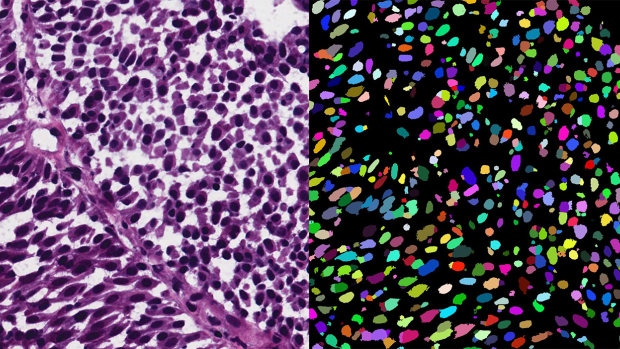
Researchers applied a new segmentation network, trained by point annotations and synthetically generated image-segmentation pairs, to automatically segment a real microscopy image (left) into its sought objects (right).
In contemporary deep learning-based methods for segmenting microscopic images, there's a heavy reliance on extensive training data that requires detailed annotations. This process is both expensive and labor-intensive. An alternative approach involves using simpler annotations, such as marking the center points of objects. While not as detailed, these point annotations still provide valuable information for image analysis.
In this study, researchers from NYU Tandon and University Hospital Bonn in Germany assume that only point annotations are available for training and present a novel method for segmenting microscopic images using artificially generated training data. Their framework consists of three main stages:
1. Pseudo Dense Mask Generation: This step takes the point annotations and creates synthetic, detailed masks that are constrained by shape information.
2. Realistic Image Generation: An advanced generative model, trained in a unique way, transforms these synthetic masks into highly realistic microscopic images while maintaining consistency in object appearance.
3. Specialized Model Training: The synthetic masks and generated images are combined to create a dataset used to train a specialized model for image segmentation.
The research was led by Guido Gerig, Institute Professor of Computer Science and Engineering and Biomedical Engineering, alongside PhD students Shijie Li and Mengwei Ren, as well as Thomas Ach at University Hospital Bonn. The three NYU Tandon researchers are also members of the Visualization and Data Analytics (VIDA) Research Center.
The researchers tested their method on a publicly available dataset and found that their approach produced more diverse and realistic images compared to conventional methods, all while maintaining a strong connection between the input annotations and the generated images. Importantly, when compared to models trained using other methods, their models, trained on synthetic data, outperformed them significantly. Moreover, their framework achieved results on par with models trained using labor-intensive, highly detailed annotations.
This research highlights the potential of using simplified annotations and synthetic data to streamline the process of segmenting microscopic images, potentially reducing the need for extensive manual annotation efforts. The research, in collaboration with the Ophthalmology department at University Hospital Bonn, is a first step in a collaboration to finally process three dimensional retinal cell images of the human eye from subjects diagnosed for age-related macular degeneration (AMD), a leading cause of vision loss in older adults.
The code for this method is publicly available for further exploration and implementation.
“Microscopy Image Segmentation via Point and Shape Regularized Data Synthesis.” S Li, M Ren, T Ach, G Gerig. arXiv preprint, arXiv:2308.09835, 2023.





