An Aspiring Doctoral Candidate Is Already Making Waves
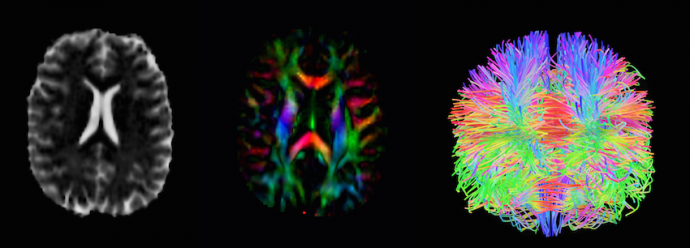
Graduate student, Neel Dey, works with High-Angular Resolution Diffusion Imaging (HARDI) as displayed in these images of an infant's brain
Neel Dey arrived in Brooklyn in 2015 with a freshly minted B.E. in Electronics and Telecommunications from the University of Mumbai and a desire to pursue a master’s degree in Tandon’s Department of Electrical and Computer Engineering. Little did he know, he says, that his research would, at times, involve shoveling quartz crystals and oil during a tunnel modelling experiment. With a deep interest in image analysis and computer vision, Dey had quickly been recruited to collaborate with a doctoral student from the Department of Civil and Urban Engineering to model particle flow in tunnels with failing structures, developing and implementing problem-specific strategies to track the exact particle displacement of individual crystals under varying pressures. In the course of that project, he fortuitously met and sought computer-vision advice from Institute Professor Guido Gerig, the chair of the Department of Computer Science and Engineering. Gerig is widely celebrated for his important work in the field of medical image analysis — a major interest of Dey’s.

Neel Dey
Dey was soon hired as a graduate research assistant in Gerig’s Visualization, Imaging and Data Analysis (VIDA) Lab, where he embarked on a joint project with Christine Curcio from the University of Alabama at Birmingham, Roland Theodore Smith from NYU Ophthalmology, Thomas Ach from University Hospital Würzburg, and others to investigate age-related macular degeneration, the leading cause of blindness in the U.S. for people over 50. “The substances that damage the macula in the human retina are unknown chemically, and therefore targeted drug treatment and early detection remains elusive,” he explains. “Biologists usually isolate materials and identify their chemical structure by putting them through a mass spectrometer. However, this hasn’t been done with drusen [the yellow deposits that are among the first signs of the condition] for various reasons, and our research group aims to do this using a machine learning approach.” In May 2017, Dey’s work will appear in four publications accepted to the Association for Research in Vision and Ophthalmology (ARVO) conference.
He will, in all likelihood, be doing so as a doctoral candidate in the Department of Computer Science and Engineering. “I’d like to bring the optimization, signal processing, and image analysis background that I have from my electrical engineering degree into the field of computer science research applied to neuroscience,” he says. “Image analysis is a field that’s hard to confine to one department: it involves electrical engineering, computer science, bioengineering, and math. Along the way, you’re equipped with an eclectic set of skills, and luckily, I’m getting the chance to apply them to some of the most relevant challenges in medical imaging.” Most recently, Dey (who, in addition to his own study and research, serves as a mentor to incoming students interested in signal and image processing) has been working with High-Angular Resolution Diffusion Imaging (HARDI) data. HARDI data can provide incredibly detailed models of nerve fibers and white matter tracts within the brain, but acquiring it is a lengthy process, and patients (particularly young ones) can rarely remain still for the duration. The VIDA Lab received, for example, a trove of elaborate datasets of infant brain scans from the University of North Carolina and the IBIS network in which more than half the diffusion gradients were unusable due to head movement. Dey, using a complex framework called HARDIPrep being developed by Shireen Elhabian at the University of Utah, was able to recover and process the data, however, and is in the process of creating and analyzing detailed models of the nerve fibers within developing infant brains. He plans, in the future, to collaborate on further developing and contributing to this emerging field of neuroimaging.




