Machine Learning for Modeling the Dynamics of the Tumor Microenvironment
Speaker:
Elham Azizi, PhD
Herbert & Florence Irving Assistant Professor of Cancer Data Research
Irving Institute for Cancer Dynamics
Assistant Professor, Department of Biomedical Engineering
Columbia University in the City of New York
Abstract:
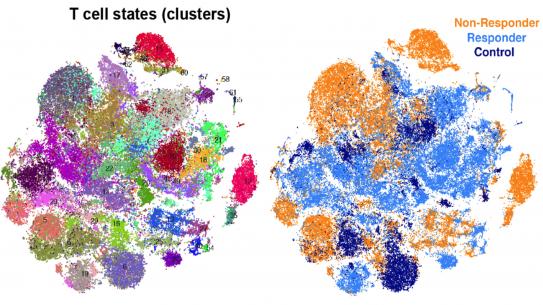
Cancer treatments lead to favorable outcomes only in a subset of patients partly due to significant heterogeneity in tumor cells. Towards developing personalized treatments tailored to each patient, Dr. Azizi’s research aims to characterize the heterogeneous populations of interacting cells in the tumor microenvironment and their dysregulated circuitry with an unsupervised approach. Recent advances in single-cell genomic technologies have empowered us to profile heterogeneous phenotypes at the resolution of individual cells. However, analysis of single-cell data involves major statistical challenges. The team of Dr. Azizi is developing novel machine learning methods to address the statistical and computational challenges inherent to high-dimensional single-cell data.
By integrating single-cell and multi-omics data using probabilistic modeling and deep learning approaches, they aim to infer dysregulated programs driving cancer stem cells as well as the reprogramming of immune cells leading to immune dysfunction. They are also interested in developing statistical models for studying the temporal dynamics of cell populations and their underlying circuitry in response or resistance to immunotherapies. Ultimately, these interpretable frameworks applied to various cancer systems can guide and improve therapies by accounting for diversity and plasticity of cells.
Dr. Azizi holds a BSc in Electrical Engineering from Sharif University of Technology (2008), and an MSc in Electrical Engineering (2010) and a PhD in Bioinformatics (2014) from Boston University. She was a postdoctoral fellow at Columbia University and Memorial Sloan Kettering Cancer Center (2014-2019). She joined the faculty of Columbia Biomedical Engineering and Irving Institute of Cancer Dynamics in 2020, where she heads the Computational Cancer Biology Laboratory.