ARISE Program Details
STEM Research Opportunity for 10th and 11th Graders

Update:
STOP: Before applying, make sure to fully read through NYU’s policies and COVID 19 safety protocols. The Center for K12 STEM follows the University's safety protocols and vaccination requirements for COVID-19 throughout all programs.
ARISE will be offered in-person for summer 2024. We will remain flexible in our response to developments related to COVID-19 and plans can of course change. The health and safety of students, faculty, staff, and our visitors is NYU's first priority, and you can read about the University's responses and reopening. If we are unable to conduct this program on campus as planned, changes to program format will be communicated promptly.
About ARISE
The NYU Tandon School of Engineering’s Applied Research Innovations in Science and Engineering (ARISE) program has graduated over 400 high school students. This free program is for academically motivated, current 10th and 11th grade New York City students with a demonstrated interest in science, technology, engineering, and math (STEM). Download the ARISE 2023 Flyer.
This seven-week program includes: a high level, 5-week authentic research experience in participating NYU faculty labs, mentoring in that placement by a graduate or postdoctoral student, a stipend of at least $750 for completing the program, as well as two weeks of workshops, college advisement and other activities geared to preparing ARISE students for the college application process. In the workshops, students will be introduced to the field of scientific ethics, contemporary issues in scientific inquiry, data collection and analysis, research practices, lab safety, and more.
With their mentors, ARISE participants will spend the latter five weeks of the program in their labs where they will make practical, substantive contributions to the lab’s research objectives, which span engineering, life sciences, and computer and data sciences. Check out the Research Opportunities section to learn more about the research going on in the over 30 ARISE labs and the professors who oversee the work.

Students will also receive training in presentation and public speaking skills, in collaboration with ARISE’s partners at Irondale Ensemble Project, and give their research findings at the program’s concluding colloquium (archival page) to NYU faculty and graduate students, their peer ARISE participants, other academic experts, family members and friends.
Through a grant from the Pinkerton Foundation, with additional support from the Depository Trust and Clearing Corporation (DTCC), ARISE is intended to provide an advanced STEM (Science, Technology, Engineering and Math) research opportunity to New York City students lacking access to high-quality STEM education experiences. Students from demographic groups underrepresented in STEM disciplines and careers, including women, students of color and those from low–income backgrounds are strongly encouraged to apply. NYU Tandon‘s ARISE program is a partner of the New York City Science Research Mentoring Consortium, a group of NYC academic, research and cultural institutions providing NYC high school students with mentored, authentic research experiences in STEM.
Research Opportunities
Please note: if you submit an application it is to the ARISE program, NOT to a specific lab. Applicants express interest in a general area of research (noted in each lab's entry below). Those students that move through the process after applying, will have the opportunity to focus on placements in a specific set of labs. However, even if you are accepted, students are not guaranteed a placement in their first-choice lab.
The following are some of the NYU labs that participate in ARISE, organized by academic department. More labs may be added in the future.
Subject Areas
AnthropologyEvolutionary Morphology Lab
Area of STEM Research: Life Sciences
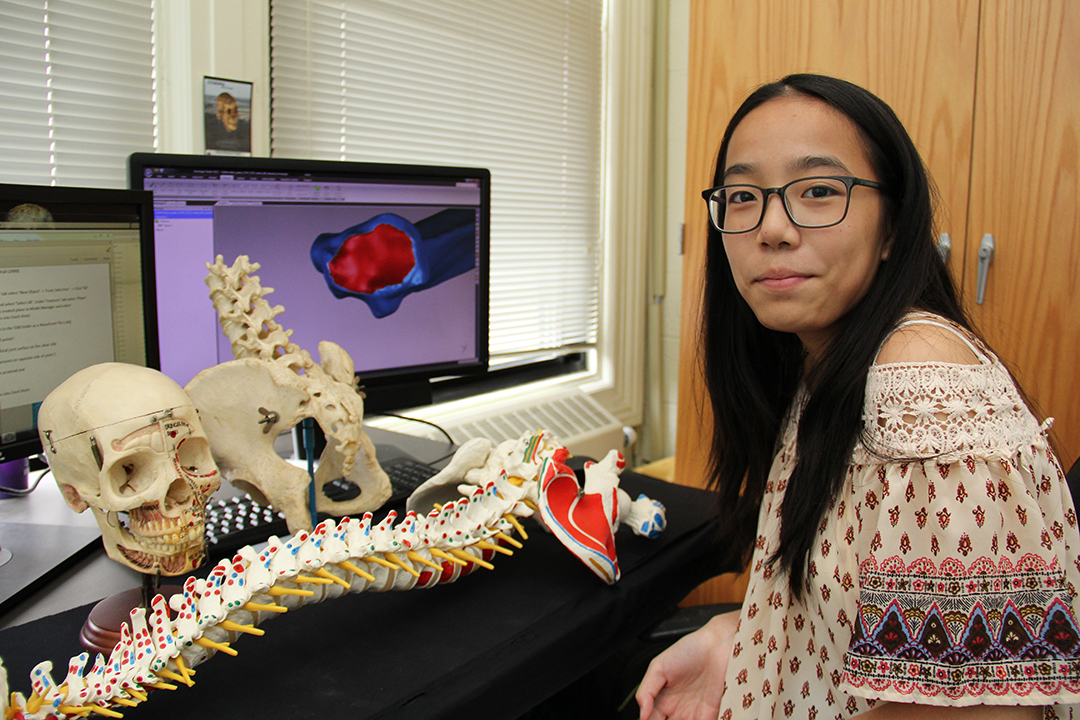
Dr. Scott Williams’ Evolutionary Morphology Lab in the Department of Anthropology at NYU focuses on lvisualization and quantification of bony morphology. In particular, Dr. Williams and his students study the skeletons of living primates in order to understand how fossil primates behaved and interacted with their environments. We use laser surface and computed tomography scanning technologies as well as traditional morphometric techniques to measure bones and fossils and compare them to known animals. Our focus is on human evolution, so much of what we dedicate our studies to are the fossilized remains of hominins — members of the human lineage that are now extinct — along with modern humans and living apes, including gibbons, orangutans, gorillas, and chimpanzees. Lab members have been involved in the study of two newly discovered species of early hominin, Australopithecus sediba and Homo naledi.
Primate Hormones and Behavior (PHaB) Lab
Area of STEM Research: Life Sciences

Dr. James Higham‘s Primate Hormones and Behavior (PHaB) Lab, an enzyme immuno-assay laboratory, measures primate hormonal and immunological analytes, often measured as metabolites from excreta (feces and urine). The lab’s overall goal is to study genetical, morphological, physiological and behavioral aspects of primate reproductive strategies as shaped by sexual selection. Following Darwin, the research is structured around two related processes: the ways in which individuals compete with members of the same sex both directly and indirectly over reproductive opportunities (intra-sexual selection); and the ways in which individuals attract members of the opposite sex (inter-sexual selection). The methods used in the lab include techniques from ethology, physical anthropology, evolutionary biology, computer vision and machine learning, experimental and comparative psychology, endocrinology and immunology, and quantitative and functional genetics.
Biology
Developmental Genomics Lab
Area of STEM Research: Life Sciences

Dr. Christine Rushlow’s Developmental Genomics Lab studies how an organism develops from a fertilized egg into an adult. We use the fruit fly, Drosophila melanogaster, as a model system because flies have a short generation time, they are easy to keep in bottles in the lab, and we can do many types of experiments with them. We study mutant flies to figure out how fly embryos develop. For example, some mutants do not form nervous systems, and thus they cannot move or eat, and they die. We ask questions like — what went wrong in that mutant during its development to give such a dramatic defect? We identify the mutant gene responsible for the defect, and then study how the gene normally works during development. We study the protein the gene makes, where is it active, and how it interacts with other proteins to produce a nervous system with the right number of nerves in the right place. Currently we are working on a gene called Zelda, which is a master regulator of genes necessary for many different processes in the young embryo, including nervous system development. Our goal is to identify all the genes Zelda regulates, and determine how they work together in a network to make an embryo with all its proper structures. ARISE students will perform many different techniques including how to synthesize RNA from DNA, prepare fly embryos for hybridization experiments, and examine embryos under the microscope. Students will learn how to analyze and interpret data, and how to assemble their data into a report.
Chromosome Inheritance Lab
Area of STEM Research: Life Sciences

Dr. Andreas Hochwagen’s research aims to discover the genetic pathways that enable sexually reproducing organisms to pass their genes on to the next generation. This research is performed in the single-celled baker’s yeast to take advantage of the fast growth and comparatively simple genome organization of this organism. Students will gain exposure to state-of-the-art methods of genetics research and will learn to perform fluorescence microscopy of chromosomes.
Molecular and Cellular Biology Lab
Area of STEM Research: Life Sciences

Dr. Fei Li and his colleagues are interested in epigenetics, a field studying heritable changes in phenotype that occur without changes in DNA sequence. Epigenetic information, stored in the form of histone modifications and DNA methylation, constitute a second layer of regulatory information important for many cellular processes, such as gene expression regulation, chromatin organization, and genome stability. We are particularly focusing on epigenetic regulation of heterochromatin and centromere in the model eukaryotic organism fission yeast (Schizosaccharomyces pombe). We take advantage of powerful experimental approaches available in fission yeast, including genetics, cell biology, biochemistry and genomics, to understand the fundamental principles of epigenetic regulation.
Chromatin Genomics Lab
Area of STEM Research: Life Sciences

In humans and other eukaryotes, genomic DNA is tightly packaged to fit inside a small nucleus. This packaging is accomplished by a set of evolutionarily conserved proteins that bind to and form a protein-DNA structure called chromatin. Dr. Sevinc Ercan’s lab studies how chromatin structure regulates gene expression. They focus on a particular family of protein complexes called condensins, which are essential for chromosome condensation in all species examined so far. To understand condensin mechanisms, Ercan lab primarily uses C. elegans, a small nematode worm as an experimental model. Students will be integrated into an ongoing project in the lab, which typically involves molecular cloning, genetics and genome editing in C. elegans.
Biomedical Engineering
Applied Micro-Bioengineering Lab

Dr. Weiqiang Chen’s Applied Micro-Bioengineering Lab focuses on development of innovative micro/nanoscale technologies and integrated biosystems targeting important emerging areas of research at the forefront of engineering and medicine. Specific examples include engineering microfluidic lab-on-a-chip systems for single-cell sensing and immune engineering, biomanufacturing organotypic organ-on-a-chip systems for cancer diagnosis and modeling, as well as biomaterials for tissue engineering and stem cell-based regenerative medicine. These novel micro/nanoengineered biomaterials and biosystems will not only permit advances in engineering but also greatly contribute to improving human health.
Chemical and Biomolecular Engineering
Protein Engineering & Molecular Design Lab
Areas of STEM Research: Life Sciences / Engineering

Dr. Jin Montclare’s Protein Engineering and Molecular Lab exploits nature’s biosynthetic machinery and evolutionary mechanisms to design new biomaterials and functional proteins. Protein engineering encompasses the optimization of biomaterials for a wide variety of applications. Of these applications, the Montclare lab uses rationale design to improve naturally-occurring proteins to become even more suitable for drug delivery, tissue engineering, plastic degradation, protein-based hydrogels and polymer materials, gene delivery, and many others.
Biomolecular Engineering Lab
Areas of STEM Research: Life Sciences / Engineering
The goal of research in Dr. Jin Ryoun Kim’s group is engineering of proteins to solve modern scientific and engineering problems related to protein stability, misfolding, aggregation, and self-assembly. Students will have research opportunities to study proteins with specific focuses on (1) understanding and modulation of protein aggregation implicated in amyloid diseases, such as Alzheimer’s and Parkinson’s diseases, and/or (2) stabilization of biocatalytic enzymes for industrial applications.
Bio-interfacial Engineering and Diagnostics Group
Areas of STEM Research: Life Sciences / Engineering

Dr. Rastislav Levicky’s Bio-interfacial Engineering and Diagnostics Group studies biological interactions. Biomolecular interactions are highly challenging to detect, yet are crucially important for identifying toxicity, medical usefulness, origins of biological material such as from viruses or harmful bacteria, and understanding how the subroutines encoded in DNA get translated to the functioning of living cells. Research in the Levicky group develops devices to measure these interactions. Potential applications range from identification of contaminants in water and food to medical treatments tailored to a patient’s genetic uniqueness, and to discovery of molecular mechanisms behind diseases such as cancer. Students will learn how to track behavior of biologically-important molecules, perform computer-based analysis of data, work to develop better hardware to analyze molecules inside of very small nanoliter volumes, and more.
Crystal Engineering Laboratory
Areas of STEM Research: Engineering and Materials Chemistry
Dr. Stephanie Lee's is investigating sustainable alternatives to silicon solar panels for renewable energy harvesting. We are replacing silicon with flexible, lightweight materials that can be formulated as liquid inks and spread across surfaces. When dried, these materials form films that convert sunlight into electricity. We are engineers of the ink-to-film transition, guiding ink molecules into three-dimensional networks of wires, plates, and cubes in the few seconds it takes for the films to dry. By controlling the size, shape, and orientation of these nanoscale structures, invisible to all but the most powerful microscopes, we are improving the light-harvesting efficiency and stability of these films so that they can compete with commercial silicon panels.
Hartman Lab
Areas of STEM Research: Engineering

Dr. Ryan Hartman’s laboratory investigates flow chemistry with microsystems using catalysis and reaction engineering principles. Our contributions impact the design of processes and systems, ranging from the molecular-to-the-macro- length scales. Continuous-flow manufacturing, flow chemistry, microchemical systems, and molecular management are major themes of our laboratory. Applied mathematics are essential in each area in order to derive predictive models that impact society. The ARISE program provides K-12 students the opportunity to engage in research related to the above-mentioned themes.
Electrochemical Engineering Lab
Area of STEM Research: Engineering
Dr. Miguel Modestino’s Electrochemical Engineering Lab investigates materials that use renewable energy sources such as wind and solar to drive chemical reactions. These types of materials are often found in widely used energy storage and conversion technologies such as batteries and fuel cells. Students involved in projects will be exposed to cutting-edge energy research tools, will be able to design and develop new prototypes of energy reactors using 3D printers, and will learn how chemical engineers can accelerate the transition towards a clean-energy world.
Nanostructured Hybrid Materials
Area of STEM Research: Engineering

Dr. Ayaskanta Sahu’s laboratory focus on designing materials at the nanoscale with dimensions that range from a few nanometers to tens of nanometers. To put these numbers into perspective, a typical nanocrystal will be around 10,000 times smaller than the diameter of human hair. While nanometer-sized chips are ubiquitous in the semiconductor industry, our lab is focused on expanding the application of these tiny nanocrystals in fields such as lighting (LEDs), solar cells, and thermoelectrics. Thermoelectrics are electronic devices that can convert waste heat into electricity. With about 60% of all usable power lost as waste heat, thermoelectrics have the potential to radically modify the current clean energy landscape. Students working in our lab will have hands-on research experience of synthesizing these nanomaterials and assembling them into working devices while learning basic concepts of chemistry, physics, materials science, and engineering.
Pinkerton Research Group
Areas of STEM Research: Engineering
Based in NYU’s Chemical and Biomolecular Engineering Department, Prof. Nathalie Pinkerton's research group focuses on developing responsive soft materials for bio-applications ranging from controlled drug delivery to vaccines to medical imaging. Taking an interdisciplinary approach, we use tools from chemical and materials engineering, nanotechnology, chemistry, and biology to create soft materials via scalable synthetic processes. We focus on understanding how process parameters control the final material properties, and in turn, how the material behaves in biological systems.
Transformative Materials and Devices (TMD) Laboratory
Area of STEM Research: Engineering

Prof. André D Taylor’s group focuses on the development of novel materials and devices for energy conversion and storage. True to their name, the group designs and studies transformative technologies that have the ability to change the status quo and promote the adoption of sustainable energy generation and use. In particular, they study next-generation lithium-ion batteries, solar cells made from easily-processed materials, along with fuel cells, devices that produce energy from combining hydrogen and oxygen into water.
Civil and Urban Engineering
Soil Mechanics Lab
Area of STEM Research: Engineering
Dr. Magued Iskander’s Soil Mechanics Lab will engage students in research on modeling soil structure-interaction using transparent soils, lasers, and digital image correlation. This research will expose students to the state of the art experimental methods used in civil engineering as well as to the advanced analytical methods that are employed for image analysis.
Urban Modeling Group Lab
Area of STEM Research: Computer and Data Science/Engineering

Dr. Debra Laefer’s Urban Modeling Group focuses on the data-driven study of the urban environment anchored in high-resolution aerial laser scanning data. Students will be engaged in a collaboration with the community in Sunset Park, Brooklyn, to maintain a digital twin of the neighborhood. Potential application areas include: urban accessibility, community-driven digital twin, public space maintenance, municipal service delivery, underground infrastructure awareness, and heritage conservation, as well use of laser scanning for 3D visualization and exploration.
Building Informatics and Visualization Lab (biLAB)
Area of STEM Research: Computer and Data Science/Engineering
Dr. Semiha Ergan’s Building Informatics and Visualization Lab (biLAB) focuses on how data can inform the design, construction, and operation of built environments and infrastructure systems in urban settings. The research team takes advantage of advancements in technology, such as building information modeling (BIM), virtual reality (VR), and machine learning, to develop information repositories, visualization platforms, and control systems that empower engineers, building owners, and facility operators to better construct, maintain, and operate buildings and infrastructure.
Behavioral Urban Informatics, Logistics, and Transport (BUILT) Lab
Area of STEM Research: Computer and Data Science/Engineering

Part of the C2SMART University Transportation Center, Dr. Joseph Chow’s Behavioral Urban Informatics, Logistics, and Transport (BUILT) lab is looking to involve students in research in transportation systems and urban mobility. Students will be exposed to computational models to evaluate networks of transportation infrastructure, routing and pricing algorithms of transportation agencies and private service operators (e.g. Uber, Lyft), and simulations of traffic and transit operations under large-scale incident scenarios. Students will help test and integrate newly acquired state-of-the-art equipment for the C2SMART Center — tablets, wearable sensors, instrumented vehicles, commercial drones, VR devices, and GIS-based video wall interfaces.
Urban Mobility and Intelligent Transportation Systems (UrbanMITS) Laboratory
Area of STEM Research: Computer and Data Science/Engineering

Part of the C2SMART University Transportation Center, The UrbanMITS Lab led by Dr. Kaan Ozbay is a multi-modal transportation infrastructure research and education facility combining a series of new concepts, technologies, and services to integrate information, vehicles, and transportation infrastructure to increase mobility, safety and comfort, and reduce energy waste and pollution. Students will be engaged in activities ranging from data analysis, coding, and modeling to building and testing sensors and equipment on projects ranging from traffic and pedestrian simulations to emergency evacuations and traveler safety. All of the work conducted in the UrbanMITS lab uses real-world data and the solutions found will be used for real-world implementation by agencies and policy-makers.
Silverman Lab (Environmental Engineering)
Area of STEM Research: Engineering and Life Sciences
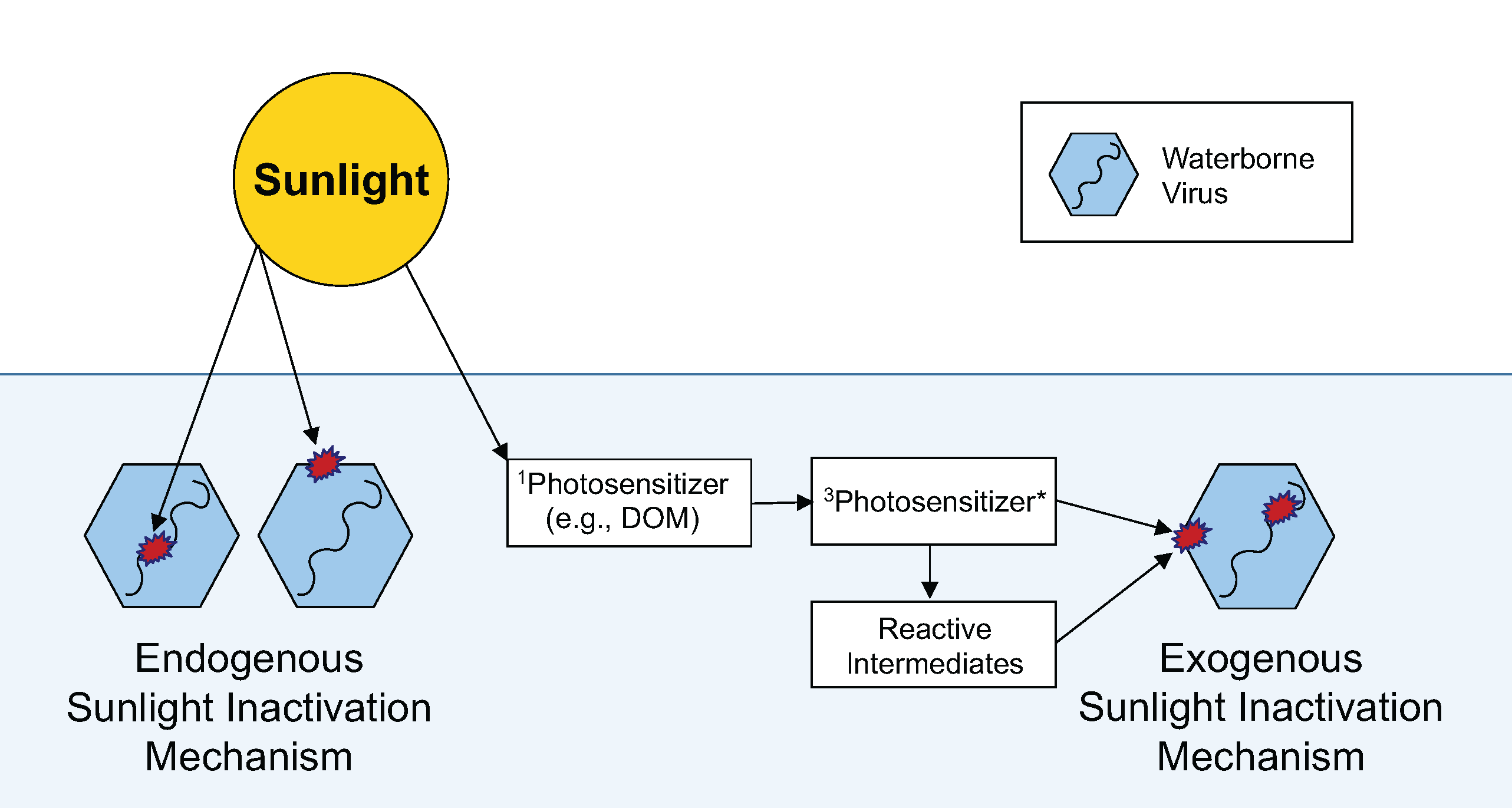
Dr. Andrea Silverman's environmental engineering laboratory conducts research to understand and design sustainable and appropriate wastewater treatment process, in an effort to protect public health and environmental quality. Our research is focused on water quality, wastewater treatment, detection and control of waterborne pathogens, design of natural wastewater treatment systems (e.g., treatment ponds and constructed wetlands), and the safe reuse of human waste. Research this summer will involve the study of the impact of algae growth on pathogen disinfection in natural systems.
FloodNet
Area of STEM Research: Engineering and Life Sciences
FloodNet, facilitated by Prof. Charlie Mydlarz, is a cooperative of communities, researchers, and New York City government agencies working to better understand the frequency, severity, and impacts of flooding in New York City. The data and knowledge gained can be used by local residents, researchers, city agencies, and others to advocate around and work to reduce flood risk. To do this we are designing, building and deploying low-cost sensors to measure street-level flooding in NYC.
Computer Science and Engineering
Area of STEM Research: Computer and Data Science/ Engineering
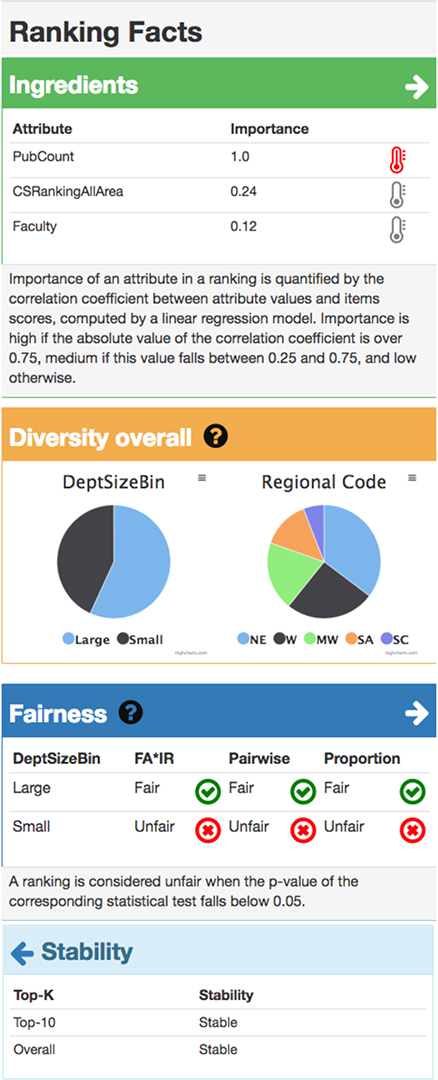
Members of Professor Julia Stoyanovich's Responsible Data Science Lab, part of the Visualization and Data Analytics Research Center at NYU, work on incorporating ethical norms and legal requirements into all stages of the data science lifecycle. Ongoing projects include developing ranking methods that produce fair, diverse, and stable results; explaining data and models with "nutritional labels"; negotiating the privacy / transparency interface with the help of privacy-preserving synthetic data generation mechanisms; and helping determine winners in elections in cases where votes are incomplete or uncertain. Students working in the lab will help develop tools and collect data related to our current projects.
Privacy and Security Automation Lab (PSAL)
Area of STEM Research: Computer and Data Science/ Engineering

Professor Rachel Greenstadt's Privacy and Security Automation Lab (PSAL) conducts research at the intersection of the privacy and security domains with machine learning. Research topics include detecting the author of given samples of source code or binary files. In order to accomplish this goal, a list of candidate authors is used to train the machine learning algorithm. Based on their interests and preferences, ARISE students will collect datasets, and later analyze them to extract useful features. Other topics of PSAL research include understanding Tor users behavior on Wikipedia edits, and analyzing the privacy concerns of systems that detect mental illness of Twitter Users.
Area of STEM Research: Computer Science/ Engineering
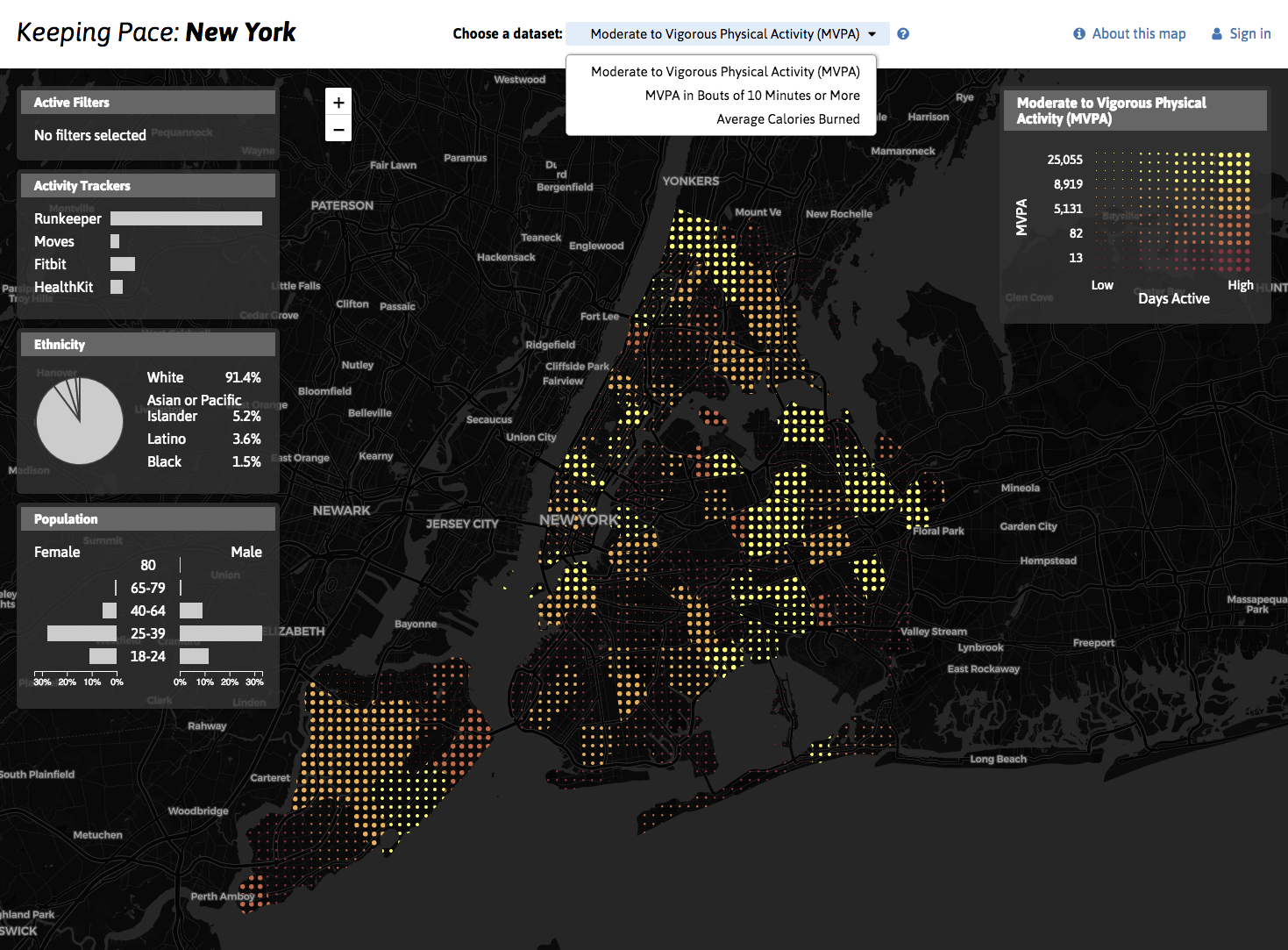 Professor Rumi Chunara's laboratory works on population-level public health problems by using Internet and mobile data sources that capture aspects of our every-day lives. This includes data such as from social media, wearables, text messaging, smartphone reports and satellite imagery. Our research involves natural language processing, machine learning and deep learning to identify relevant features from such data, and then incorporating them into statistical models of disease over space and time. We work on many types of public health issues including predicting infectious disease events and measuring social and other risk factors that can also affect our health, like substance or even discrimination.
Professor Rumi Chunara's laboratory works on population-level public health problems by using Internet and mobile data sources that capture aspects of our every-day lives. This includes data such as from social media, wearables, text messaging, smartphone reports and satellite imagery. Our research involves natural language processing, machine learning and deep learning to identify relevant features from such data, and then incorporating them into statistical models of disease over space and time. We work on many types of public health issues including predicting infectious disease events and measuring social and other risk factors that can also affect our health, like substance or even discrimination.
Visualization and Data Analytics Research Center
Area of STEM Research: Computer and Data Science/ Engineering
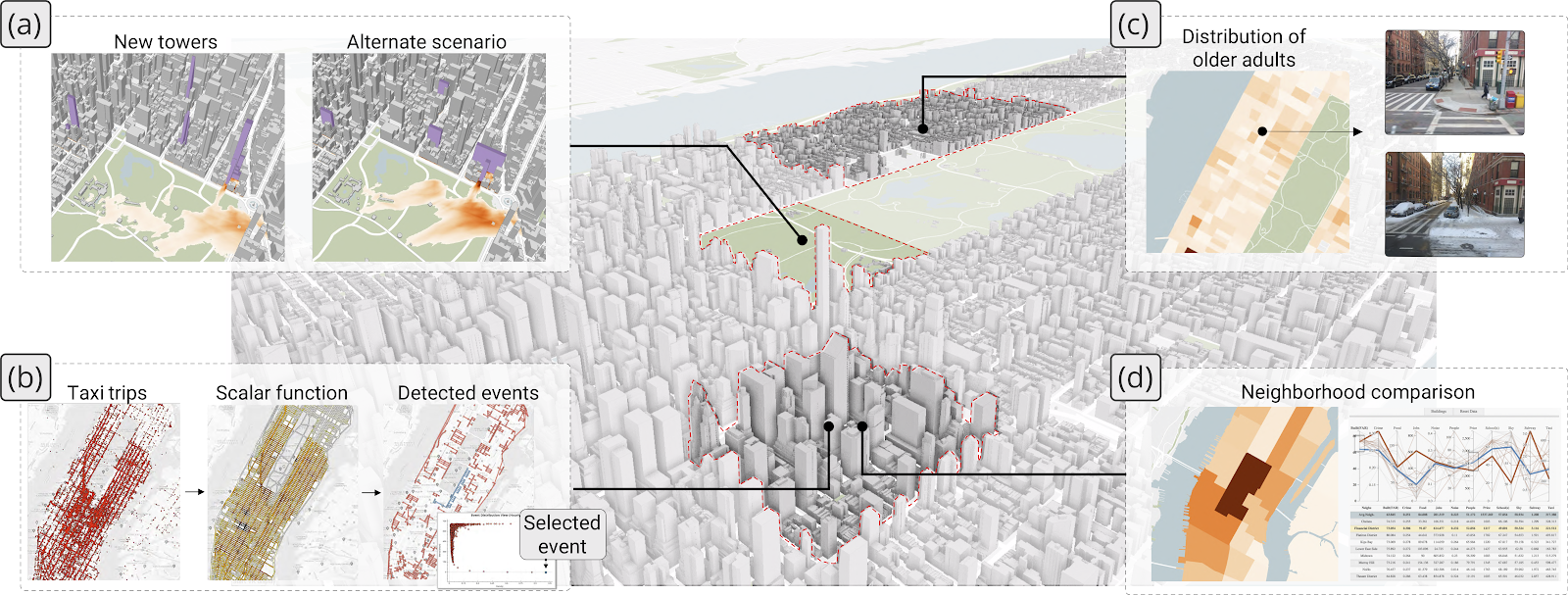
The NYU Visualization, Data Analysis and Imaging Research Center, co-led by Professor Juliana Freire and Professor Claudio Silva, brings together an interdisciplinary group of computer scientists who work closely with domain experts to address real, societally important problems. We develop methods and systems that enable a wide range of users to obtain trustworthy insights from data. This spans topics in large-scale data analysis and integration, visualization, machine learning, provenance management, and web information discovery, and different application areas, including urban analytics, sports analytics, predictive modeling, and computational reproducibility. Ongoing projects include the development of methods to support data discovery and dataset search, analytics to empower experts to explore complex relationships between spatio-temporal datasets, development of new techniques (including hardware) for sports data acquisition, analysis and visualization of sports data (including e-sports), and automated model discovery systems that enable users with subject matter expertise but no data science background to create effective machine models. ARISE students working at the lab will do research related to these projects based on their preferences and interests.
Machine Learning for Good Laboratory (ML4G)
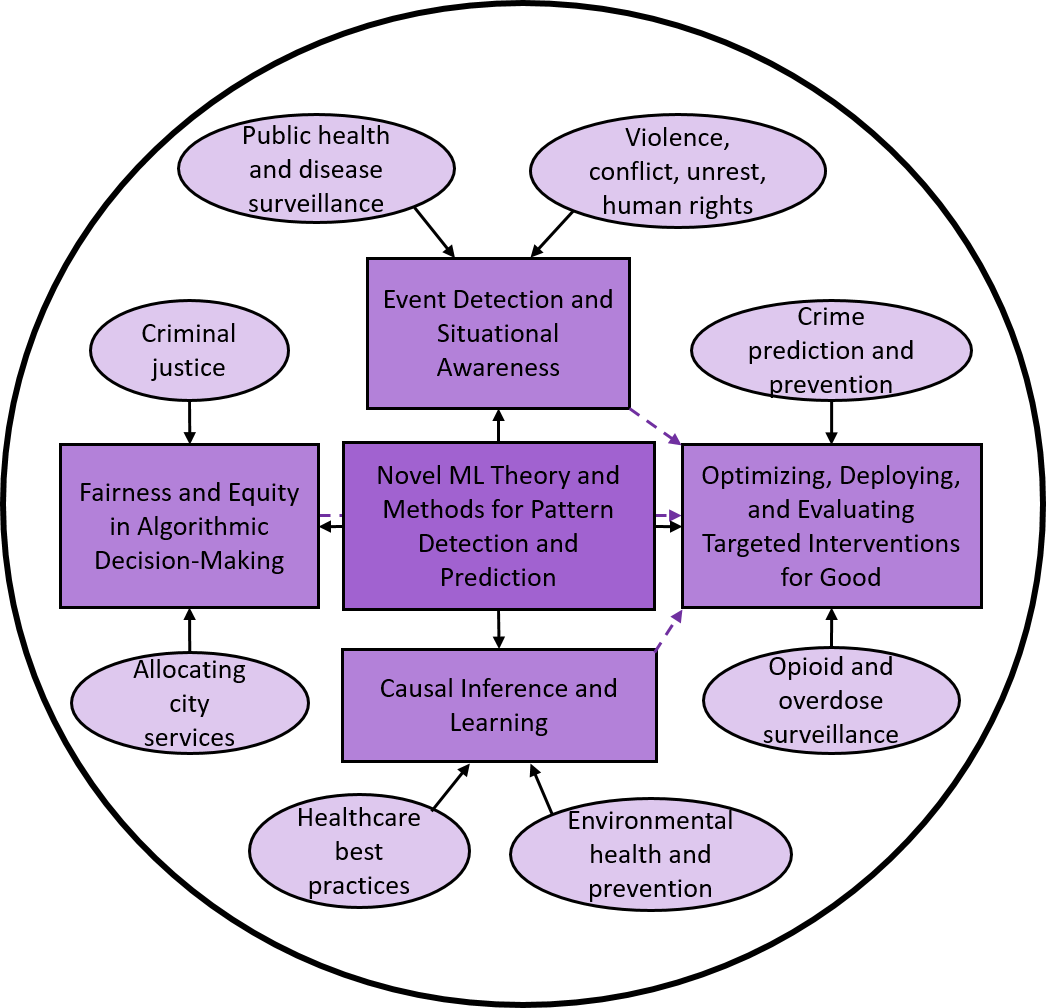
Area of STEM Research: Computer and Data Science/ Engineering
NYU's Machine Learning for Good Laboratory, directed by Prof. Daniel B. Neill, is focused on developing new machine learning methods and applying them for social good. For example, we have developed methods to detect emerging disease outbreaks earlier and more accurately, to target public health interventions more effectively and more equitably in response to the opioid overdose crisis, and to improve the fairness of prosecutorial decisions in criminal justice. ARISE students working with the ML4G Lab will join our NSF-funded Fairness in Artificial Intelligence project, which focuses on assessing and improving fairness in decision making (including decisions made by humans, by machines, and the two working together), together with public-sector collaborators in the criminal justice, housing, and health domains. Specific tools to be developed, and application areas, can be determined based on student preferences and project needs.
Area of STEM Research: Computer Graphics and Engineering, VR/AR
The Immersive Computing Lab, directed by Prof. Qi Sun, conducts cutting edge research that spans the fields of computer graphics, physics, and computational cognition, with the goal of creating unprecedented virtual and augmented reality systems to revolutionize urban life. Our research directions include novel multimodal/low-latency/immersive interaction devices, bio-physically inspired displays, perception-aware VR/AR/Computer Graphics, and beyond.
Electrical and Computer Engineering
Center for Advanced Technology in Telecommunications (CATT)
Areas of STEM Research: Computer and Data Science / Engineering

Dr. Shivendra Panwar’s lab focuses on developing the next generation of Internet technologies. Research topics include wireless networks, telecommunications policy, video delivery over networks, network security, and more. Students work with research testbeds to implement large-scale network experiments and evaluate new applications, protocols, and ideas to make the Internet work better for everyone.
Music and Audio Research Lab (MARL)
Areas of STEM Research: Computer and Data Science / Engineering

Professor Juan Pablo Bello leads the Machine Listening group of the Music and Audio Research Lab (MARL), which focuses on endowing computers with listening skills, such as the ability to automatically identify sound sources in urban and natural environments. The team works in a diverse set of applications at the intersection between machine learning, audio signal processing and citizen science. As an example, in one of our main projects, Sounds of New York City, we develop methods to effectively monitor, analyze, and mitigate urban noise pollution. Student researchers will contribute to building, testing, and deploying remote acoustic sensors for urban noise monitoring; developing tools for mapping bird migration; and writing software for analyzing environmental sounds and animal vocalizations.
ECE Machine Learning Lab
Area of STEM Research: Computer and Data Science/ Engineering

Founded by Professor Anna Choromanska, the Machine Learning Lab at NYU Tandon School of Engineering aims to create a favorable workspace to drive innovation in the field of artificial intelligence. The members of the lab pursue research at the intersection of machine learning, deep learning, and robotics. In addition, several other faculties members inside and outside the department with specific interests in machine learning have collaborations on various projects at the lab.
Some of the ongoing projects include generative modeling and sensor fusion for self-driving vehicles, interpretability and visualization of deep learning models, multi-label classification to tag data with classes from extremely large sets of labels and designing optimization algorithms for fast and efficient training of deep learning models.
Laboratory for Nanoelectronics Research
Areas of STEM Research: Computer and Data Science / Engineering
The NYU Laboratory for Nanoelectronics Research, or Nanolab, led by Professor Davood Shahrjerdi, studies the physics of electronic materials and their application in building devices and circuits. We are an experimental group with experience in the synthesis of layered materials, nanofabrication of electronic devices, and electrical measurements at both room and cryogenic temperatures. Our team is currently pursuing the following projects: electronics for interfacing with living cells, to build chemical imagers for monitoring chemical activities across living cells in real-time with high spatial and temporal resolutions, and nano-enabled security, focusing on how to use the advantageous features of nanomaterials for secure computing and communications.
Mechanical and Aerospace Engineering
Composite Materials and Mechanics Laboratory
Area of STEM Research: Engineering
 Dr. Nikhil Gupta’s Composite Materials and Mechanics Lab will allow students to engage in research projects related to additive manufacturing (3D printing) of composite materials. The recent work is focused on applying machine learning to microstructures of composite materials for materials design and mechanical property estimation. In addition, the laboratory is working on cybersecurity in additive manufacturing to develop secure and trustworthy designs containing anti-counterfeit production features, anti-reverse engineering methods and genuine product authentication methods.
Dr. Nikhil Gupta’s Composite Materials and Mechanics Lab will allow students to engage in research projects related to additive manufacturing (3D printing) of composite materials. The recent work is focused on applying machine learning to microstructures of composite materials for materials design and mechanical property estimation. In addition, the laboratory is working on cybersecurity in additive manufacturing to develop secure and trustworthy designs containing anti-counterfeit production features, anti-reverse engineering methods and genuine product authentication methods.
Mechatronics Lab
Areas of STEM Research: Engineering / Computer and Data Science

Dr. Vikram Kapila’s Mechatronics Lab will engage students in research on the use of mobile devices to produce intuitive and natural interfaces for human robot interaction. This research has applications for children and adults who may have cognitive or physical disabilities, and it can hasten the widespread adoption of robotics in society — in homes, grocery stores, museums, etc.
Applied Dynamics & Optimization Lab
Areas of STEM Research: Robotics / Engineering / Computer and Data Science

Dr. Joo H. Kim’s Applied Dynamics and Optimization Laboratory studies multibody system dynamics, optimization theory and algorithms, motion planning, design, and control, with their applications to mechanical systems. Main domains for implementation and applications include robotic and biomechanical systems and their intersections such as lower-body wearable robots. Current research focuses on energetics of dynamic systems, legged balance and gait stability, and integration of dynamics/control with numerical optimization. Our research aims at bringing together both robotic and biomechanical systems; we aim to establish mathematical models, quantitative criteria, and algorithmic/computational foundations for their implementations in advancing: (i) design and control of robots and machines and (ii) prediction and analysis of biomechanical systems.
Dynamical Systems Lab
Areas of STEM Research: Engineering / Computer and Data Science

Dr. Maurizio Porfiri’s Dynamical Systems Laboratory conducts multidisciplinary research in the theory and applications of dynamical systems, motivated by the objectives of advancing engineering science and improving society. The research covers a wide range of topics: animal and human behavior, complex systems, control theory, dynamical systems theory, experimental and theoretical mechanics, network science, and robotics. Among the projects students may design and fabricate a robotic fish that can interact with real fish; study the spreading of epidemic in cities; understand the reasons behind gun violence; and build devices that can help blind and low-vision people to navigate and avoid obstacles.
Area of STEM Research: Robotics/ Computer and Data Science
Dr. Ludovic Righetti's Machines in Motion laboratory aims to understand the fundamental principles of robot movements: what algorithms would allow a robot with arms and legs to walk around and manipulate objects autonomously? It develops novel algorithms based on optimal control and reinforcement learning. The laboratory offers various projects such as algorithms to enable a robot to learn how to walk, optimal control experiments with a jumping quadruped robot, learning the dynamics of fast contact interactions, manipulation of complex objects and fusion of multiple sensory information to make sense of the environment.
Who Can Apply?
Applicants must meet all the criteria below:
- Live in New York City (Attending school in New York City is not sufficient).
- Completing 10th or 11th grade in June 2024.
- Commit to the entire program, full time (approximately a 9 to 5 day, Monday-Friday, everyday), from June 3 to August 9, 2024, and an orientation on May 31, 2024.
- Have a passion for science, technology, engineering and math.
- Be responsible, highly motivated students who have demonstrated timeliness, persistence, and an ability to fulfill commitments.

We know students have different coursework opportunities and may not have access to Advanced Placement courses or easy access to Regents exams. Therefore, there are no specific courses, grades, or test scores that are prerequisites. We will consider your application in its totality and will focus particular attention on your written essay. We are seeking academically motivated students with aptitude in science and math. We are looking for NYC students with a passion for STEM subjects, who are excited by the idea of conducting research and willing to commit and work hard.
If you have further questions about eligibility, please read our Frequently Asked Questions
Important Dates and Information
- Week of November 22, 2023: Application period opens online for Summer 2024.
- March 1, 2024 at 11:59 PM: Student online applications due. The web form will close after 11:59 PM EST on March 1 or Applications will remain open till spaces are filled.
- March 1, 2024 at 5 PM: Recommender online responses due.
- 2nd Week of April: Notification of applicants invited to continue with the application process by attending lab tours and group interviews at the School of Engineering or College of Arts and Sciences.
- April 16 - 19: Mandatory lab tours and group interviews.
- 3rd Week of April: Notification of applicants invited to continue with lab interviews.
- April 28 or April 29: Mandatory interviews with lab personnel and mentors.
- 2nd Week of May: Notification to selected applicants offered lab placements. All other active applicants will be waitlisted. It is very likely that several waitlisted candidates will eventually be offered lab placements.
- Within 36hrs of Notice: Students offered placements must accept or decline the offer.
- May 31 1:00PM - 4:00PM EST : Mandatory orientation for accepted students.
- June 3, 2024 to August 9, 2024: ARISE runs full-time, 5 days per week (except July 4). The final colloquium will be held on the last day, August 9, 2024.
Good to know:
- Students completing the entire program will receive a $750 stipend. Additional stipends may be available for certain students on an individual and demonstrated-need basis.
- Approximately 76-80 placements are available for summer 2024.
- We cannot accept applications from students who reside outside of one of the five NYC boroughs. Students who will be living in NYC for the summer, only, or who attend school in NYC but live outside of NYC are not eligible.
- Questions? Please read all instructions and program web pages. For information not found on the website, email: k12.stem@nyu.edu
- Follow us on social media on our Twitter, Instagram, and Facebook!

ARISE Application
Read How to Apply for detailed instructions on what is required and how the application process works.
Have all your application materials before beginning the online submission process as you can not save your progress.
Ready to Apply? Go to the Application Form.



